Webinar Info
Date: Thursday, January 28th
Time: 12:00pm ET
Presenter: Bryce Daines, VP of Product, Tute Genomics
Register Here
If your informatics infrastructure does not support phenotypic-driven variant triage, then it’s likely your NGS turnaround times are too long and your costs far too high.
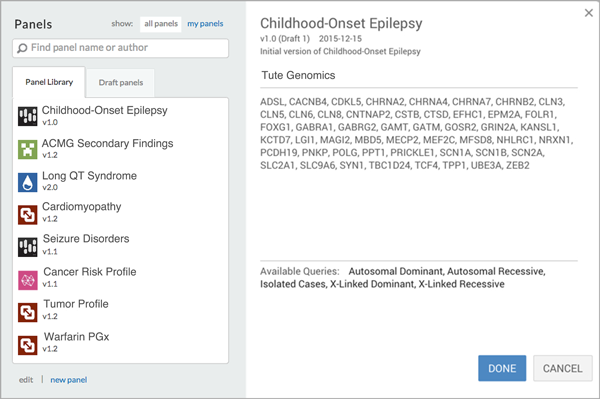
The problem is that today’s larger NGS tests produce too many genomic variants for a laboratory to review at scale. The common solution is for a provider to order a single gene test and then reflex to a larger gene panel or exome as needed. This approach is costly and time consuming.
Join us Thursday, January 28th at 12:00pm ET for the second installment of our Democratizing Regulated Clinical Sequencing webinar series where we will introduce you to an in silico panel approach designed around clinical indications that enables a laboratory to consistently run larger panels or exomes at scale, while reducing the variant review process from hours to minutes.
Learn how you can:
-
Use phenotype and inheritance mode to build a library of production-grade in silico panels that captures your organization’s expertise and mirrors your lab’s existing test menu.
-
Deploy in silico panels with one click to generate a short-list of highly relevant variants.
-
Perform “reflex” testing in real-time by adding genes on the fly without adding sequencing costs.
-
Empower labs and clinical providers to leverage up-to-date genomic reference databases to reanalyze samples as a patient’s clinical presentation evolves.
-
Catalog reviewed variants in a central repository to reduce costly rework.